Shi Zhengli and other Chinese scientists discovered that the evolutionary “arms race” shaped the diversity of viruses and their receptors. Identification of key residues involved in interspecies transmission is very important for predicting potential pathogens and understanding how viruses transition from wild animals to humans.
Previously, researchers have identified SARS-associated coronaviruses (SARSr-CoV) with different genetic characteristics in Chinese rhinoceros. This latest study also demonstrates the high diversity of the bat receptor ACE2 (Angiotensin Converting Enzyme 2) in the Chinese chrysanthemum bat population. These ACE2 variants support SARS virus and SARS-associated coronavirus infections, but have different binding affinities for different spike proteins. The SARS-associated coronavirus spike protein has a higher binding affinity for human ACE2, showing that these viruses have the ability to infect humans by transition. The positive selection of residues at the interface between ACE2 and SARS-associated coronavirus spike proteins indicates that there is long-term and sustained co-evolutionary dynamics between them. Therefore, continuous monitoring of this group of viruses in bats is necessary to prevent the next SARS-like disease.
The above research comes from a paper published on the pre-print platform bioRxiv by Shi Zhengli team of Wuhan Institute of Virology, Chinese Academy of Sciences and Professor Ouyang Songying, School of Life Science, Fujian Normal University: Evolutionary arms race between virus and host drives genetic diversity in bat SARS related coronavirus spike genes.
The Chinese rhinoceros bat is the host of the SARS virus, and it also carries a variety of SARS-related coronaviruses. These viruses have a high degree of genetic diversity, especially the spike protein genes of the virus. Despite varying degrees of variation, some bat SARS-related coronaviruses can still use the human receptor ACE2 to enter human cells. The researchers speculate that there is an interaction between the ACE2 receptor of the bat and the SARS-associated coronavirus spike protein, and this drives the genetic diversity of the SARS-associated coronavirus.
Researchers have identified a series of ACE2 variants of Chinese rhinoceros, some of which have polymorphic positions that interact with SARS-CoV spike protein point. Pseudoviruses or SARS-associated coronaviruses carrying different spike proteins have different transient infection efficiencies in cells expressing ACE2 variants of bats. By measuring SARS virus, SARS-associated coronavirus spike protein and batThe binding affinity between bat receptors and human receptor molecules can be observed in related results.
All tested bat SARS-associated coronavirus spike proteins have higher binding affinity to human ACE2 than to bat ACE2. However, the binding affinity of SARS-associated coronavirus spike protein to human ACE2 is 10 times lower than that of SARS-CoV spike protein to human ACE.
Structural modeling indicates that the difference in binding affinity between the spike and ACE2 may be due to changes in some key residues in the interface between these two molecules. Molecular evolution analysis shows that these residues are in strong positive selection.
These results indicate that the SARS new crown virus spike protein and bat ACE2 may evolve with each other over time and experience each other ’s selection pressure, thus triggering the evolution of “ The dynamics of the arms race. This further proves that the Chinese chrysanthemum bat is the natural host of SARS-associated coronavirus.
Coronaviruses are enveloped viruses that contain single-stranded positive-strand RNA. There are four genera in this subfamily, namely α, β, γ and δ. Alpha coronavirus and beta coronavirus originate from bats or rodents, while gamma coronavirus and delta coronavirus originate from birds. Since the beginning of the 21st century, three beta coronaviruses have caused severe pneumonia outbreaks in humans. They are SARS-CoV, MERS-CoV and SARS-CoV-2.
The outbreak caused by SARS-CoV-2 reminds people of the SARS outbreak that occurred 17 years ago. SARS is a zoonotic disease. In the next few years, scientists have detected or isolated 75 SARS-associated coronaviruses (SARSr-CoV) with different genetic characteristics from bats in different regions of China and Europe.
The SARS-associated coronavirus in bats has 96% nucleotide sequence similarity to the SARS-CoVs of humans and civets, and the most variable region is spike protein (S) and accessory proteins ORF3 and ORF8. In addition, the researchers have identified all the genetic building blocks of SARS-CoV in the SARS-associated coronavirus genomes of different bats, indicating that the ancestors of SARS virus were derived from the recombination of the SARS-associated coronavirus genome of bats, which originated from bat .
The first step in viral infection is to identify cell receptors, which is also an essential step. CoronavirusEntry is mediated by the specific interaction between the viral spike protein (Sike, S) and cell surface receptors, and then the fusion between the virus and the host membrane occurs. Coronavirus spike proteins are functionally divided into two subunits: cell attachment subunit (S1) and membrane fusion subunit (S2). The S1 region contains an N-terminal domain (NTD) and a C-terminal domain (CTD); both can be used for coronavirus receptor binding (RBD).
For SARS-CoV, S1-CTD acts as an RBD and binds to the cell ’s receptor, angiotensin converting enzyme 2 (ACE2). Cryo-electron microscopy and crystal structure analysis identified some key residues in the interface between SARS virus S-RBD and human ACE2.
According to the size of S protein, bat SARS-associated coronavirus can be divided into two different clades. Clade 1 contains viruses with spike proteins of the same size as SARS viruses. Due to the deletion of 5, 12, or 13 amino acids, the spike protein of clade 2 virus is smaller than that of SARS virus.
Although RBD is different, all clade 1 strains can use ACE2 to enter the cell, while clade 2 strains cannot enter directly due to the aforementioned deletion. These results indicate that, in terms of genome similarity and the use of ACE2, clade 1 members are likely to be a direct source of SARS virus.
ACE2 is functionally divided into two domains: the N-terminal domain participates in SARS-CoV binding, and the C-terminal domain participates in the regulation of cardiac function. Previous results indicate that the C-terminal domains of ACE2 from different sources are relatively conserved, while the N-terminal domains show more diversity among species. Previously, it has been proved that SARS virus can use ACE2 of water mouse ear bat and ACE2 of Chinese chrysanthemum bat. A small mutation in the RBD binding site can change ACE2 from being less susceptible to SARS-CoV binding to susceptibility. Since all SARS-associated coronaviruses belonging to Clade 1 can be extracted from the Chinese chrysanthemum, and all can also use ACE2, the researchers asked whether the mutation in ACE2 of Chinese chrysanthemum may cause SARS in bats Diversity of related coronaviruses.
The research team studied the polymorphism of the ACE2 gene of the Chinese bat head bat, and combined molecular evolution analysis, protein affinity determination and virus infection measurement to evaluate their Sensitivity and binding affinity of SARS-associated coronavirus spike proteins from different bats.
The results indicate that the spike protein diversity of SARS-associated coronavirus may be subject to the natural selection pressure of ACE2 variants of Chinese rhinoceros; during long-term coexistence, SARSr-CoV spike protein may be ACE2 is selected to maintain its own genetic diversity and is suitable for the population of A. chinensis.
The ACE2 gene is highly polymorphic in the Chinese chrysanthemum bat population
The ACE2 gene is highly polymorphic in the Chinese chrysanthemum bat population
According to the prevalence of bat SARS-associated coronavirus and the sample tissue For availability and quality, the researchers used samples from three provinces (Hubei, Guangdong, and Yunnan) for ACE2 amplification.
In addition to the bat ACE2 (sample IDs 832, 411 and 3357 collected from Hubei, Guangxi and Yunnan, respectively) and other bat ACE2 (GenBank registration number ACT66275 , This is a sample collected from Hong Kong), the researchers obtained the ACE2 gene sequence from 21 individuals of Chinese chrysanthemum bat: 5 in Hubei, 9 in Guangdong, and 7 in Yunnan. These bat ACE2 sequences show 98-100% amino acid identity within their species and 80-81% amino acid identity with human ACE2.
These bats ACE2 have observed major changes in the N-terminal region, including some residues that have previously been identified as being in contact with the S-RBD of the SARS virus. Eight residues were identified based on non-synonymous SNP analysis, including 24, 27, 31, 34, 35, 38.41 and 42. The combination of these 8 residues produced 8 alleles, including RIESEDYK, LIEFENYQ, RTESENYQ, RIKSEDYQ, QIKSEDYQ, RMTSEDYQ, EMKT KDHQ and EIKT EIKTKDHQ, respectively named alleles 1-8.
In addition to the ACE2 genotype data from previous studies (alleles 4, 7 and 8), the researchers also identified 5 in the Chinese chrysanthemum bat population New alleles. “Allele 2” was found in samples from two provinces, “Allele 4” was found in three provinces, and other alleles appeared to be geographically restricted. In summary, 3 alleles (4, 6 and 8) were found in Guangdong, 4 alleles (1, 2, 4 and 7) were found in Yunnan, and 3 alleles (2, 4) were found in Hubei And 5), 1 allele was found in Guangxi and Hong Kong respectively. In a bat cave in Yunnan where the direct ancestor of the SARS virus was found, The researchers found that 4 alleles coexist.
To sum up, these data indicate that the ACE2 variant has been in the long-term existence of the Chinese chrysanthemum bat population in different regions. The substitution of sites that directly contact the S-RBD of SARS virus indicates that they may have important functions in the evolution and spread of SARS virus.

Different sensitivities of A. chinensis ACE2 variants to SARS-associated coronavirus infection

Different sensitivities of A. chinensis ACE2 variants to SARS-associated coronavirus infection
To evaluate different ACE2 Whether the molecule will affect the entry of SARS virus and bat SARS-associated coronavirus, the researchers transiently expressed ACE2 variants of Heliotrope chinensis in HeLa cells, and tested SARS virus pseudotypes carrying different spike proteins or bat SARS The entry efficiency of coronaviruses.
4 strains of bat SARS-associated coronavirus can be divided into 4 genotypes according to S1 sequence. Simply put, compared with the spike protein of SARS virus, the RBD of SARS-associated virus RsWIV1 has a high degree of amino acid identity with SARS virus; RsWIV16 is a close relative of SARS virus, showing high amino acid similarity in NTD and RBD; Rs4231 on NTD It has high amino acid similarity with SARS virus; RsSHC014 is different from SARS virus in NTD and RBD regions.
Similar to previous results: all four bat SARS-associated coronavirus strains with the same genomic background but different spike proteins can be similarized using human ACE2 Horizontal replication.
However, there are some differences in how they use ACE2. All test viruses can effectively use alleles 1, 2, 4, and 5 to enter. RsWIV1 and RsWIV16 with the same RBDThen allele 6 from Guangdong (sample ID 1434) cannot be used. Rs4231 and RsSHC014 with the same RBD cannot use allele 7 (sample ID 3357) and allele 8 (sample ID 1438) from Yunnan and Guangdong, respectively. The SARS virus BJ01 has a high similarity to the RBD of WIV1 and WIV16, and can use the same bat ACE2 allele as Rs4231 and RsSHC014 in pseudotype infection experiments.
These results indicate that virus entry into cells is affected by the spiked RBD and ACE2 variants.
Mutations of ACE2 residues in bats will affect their binding affinity to SARS-associated coronavirus RBD

Mutations of ACE2 residues in bats will affect their binding affinity to SARS-associated coronavirus RBD
For further understanding of SARS virus and SARS-associated coronavirus pairs The ability to use ACE2 was different. The researchers expressed the RBD of the coronaviruses BJ01, RsWIV1, RsWIV1, RsSHC014, and ACE2 of three Chinese rhinoceros bats, which were allele 6 (sample 1434) and allele 7 (sample 3357) And allele 2 (sample 5720).
Subsequently, the researchers tested the affinity between them. The positive control is the RBD of SARS virus BJ01 and human ACE2, and the negative control is the RBD of SARS virus RsWIV1 and human DPP4. Real-time analysis shows that, based on the equilibrium dissociation constant KD, different RBDs have different binding affinity to ACE2.
Consistent with the results of the virus infection experiment, it was found that 1434ACE2 (allele 6) binds to RsSHC014 and BJ01, but not to the RBD of RsWIV1; the study also found 3357ACE2 (etc. Allele 7) binds to RsWIV1, but not to RBD of RsSHC014 and BJ01; 5720ACE2 (etc.Allele 2) was found to bind all tested RBDs. All RBDs tested have high binding affinity to human or bat ACE2. However, compared with the RBD of the two bat SARS-associated coronaviruses, it has a low binding affinity for ACE2.
These results show that the binding affinity of spiked RBD and ACE2 affects the virus ’s ability to infect.
Structural model of the interaction between the SARS-associated coronavirus RBDs and the Chinese rhinoceros ACE2s

Structural model of the interaction between the SARS-associated coronavirus RBDs and the Chinese rhinoceros ACE2s
The size of the spike proteins of four bat SARS-associated coronaviruses The same amino acid homology with SARS-CoV exceeds 90%, indicating that these proteins have 228 similar structures. In this study, they constructed a structurally complex model of the bat SARS-associated coronavirus RsSHC014 RBD using ACE2 3357 (allele 7), and constructed a bat SARS using ACE2 1434 (allele 6) A complex model of the coronavirus RsWIV1 RBD.
This is consistent with the results of the SARS-CoV RBD binding affinity experiment with human ACE2. Compared with the contact residues at the interface of SARS-CoV RBD and human ACE2, two viral binding hot spots (hotspot hotspot Lys31 and Lys353 consisting of Glu35 and Asp38 salt bridge respectively) on or near ACE2 can be observed. As mentioned earlier, these two hot spots are hidden in a hydrophobic environment. They provide a large amount of energy for the virus-receptor binding interaction and fill the key gaps in the hydrophobic superposition 238 interaction at the binding interface.
Compared with human ACE2, the two main residual changes of A. chinensis ACE2-3357 are E35K and Y41H. E35K and RsSHC014 RBThe breaking of the salt bridge of K31 and Arg479 in D may affect the binding affinity between them. The histidine at position 41 may weaken the support of the K353-d38 salt bridge, because histidine is less hydrophobic than tyrosine, resulting in a reduced binding affinity for BJ01 RBD. Therefore, the binding affinity of BJ01 and RsSHC014 RBD to ACE2-3357 is low.
Threonine was found at position 31 in ACE2-1434, which is different from the position of human ACE2, which is lysine. Although the change of K31T prevents it from forming a salt bridge with Glu35, Tyr442 in RBD c of BJ01 can form a hydrogen bond with it. However, the serine at 442 in RsWIV1 RBD does not have this function.
In addition, RsSHC014 contains an arginine at 479, Thr31 cannot form a salt bridge with Glu35, so that Glu35 can form a salt bridge with Arg479, but RBD residue in RsWIV1 Asn479 may cause it to lose this ability. Therefore, BJ01 and RsSHC014 RBD can bind ACE2 -1434, but RsWIV1RBD cannot bind ACE2 -1434.
All ACE2 in this study contained an asparagine at position 82, while human ACE2 is methionine at this position. The change in M82N introduces an unfavorable hydrophilic residue, weakening the hydrophobic network around hot spot 31. In addition, Asn82 introduced a glycosylation site, as in rat ACE2, which prevented it from effectively supporting SARS-CoV infection; glycan at position 82 of ACE2 may cause spatial interference with viral RBD binding.
Therefore, M82N may have a significant effect on the interaction of SARS-CoV and SARS-associated coronavirus RBD with Chinese ceratoides ACE2s. As mentioned earlier, residue 487 of RBD interacts with hot spot 353 on ACE2. RsWIV1 RBD contains Asn487, the polar side chain of Asn487 may adversely interact with the aliphatic part of Lys353 residue in ACE2, affecting the hot spot interaction between K353 and D38.
In addition, RsSHC014 RBD contains an alanine at position 487; the small side chain of Ala 487 does not support hot spot 353structure. Therefore, the binding affinity of RsWIV1 and RsSHC014 RBD to human ACE2 is lower than that of BJ01, but the binding affinity to human ACE2 is higher than ACE2, which is highly consistent with the results of virus infection and binding experiments.
The SARSr-CoV spike gene co-evolves with ACE2 of Chinese rhinoceros through positive selection

The SARSr-CoV spike gene co-evolves with ACE2 of Chinese rhinoceros through positive selection
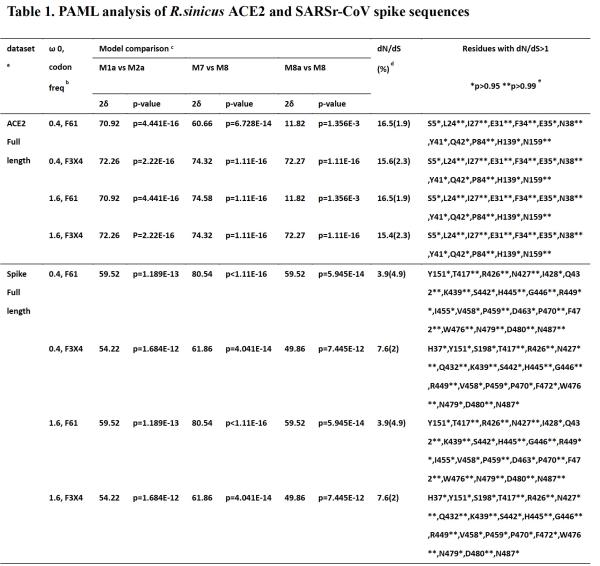
To test the spike protein and SARS virus spike protein The selection pressure of the ACE2 gene of Chinese chrysanthemum bats, they used the coding ml program of the PAML software package to analyze the ratio of non-synonymous mutations to synonymous mutations (dN / dS ratio) of a single codon. By analyzing the complete gene sequence encoding the SARS-associated coronavirus spike protein, and aligning the gene sequences of 9 SARS-associated coronavirus spike proteins from the Chinese scorpion bat samples.
The study found that the model allows codons to evolve under positive selection (M2a and M8). In model M8 (initial seed value ω (dN / dS) = 1.6, codon frequency = F3X4), 20 codons (p> 0.95) are positively selected at a rate of dN / dS> 1, according to the crystal structure, 298 17 of the codons were found in the RBD region, facing the receptor ACE2. In addition, the five codons (442, 472, 479, 480, and 487) appearing in SARS-CoV spikes have been previously determined to have a significant effect on the binding affinity of human ACE2.
Next, they analyzed the ACE2 gene of Chinese chrysanthemum chinensis by comparing the 25 ACE2 gene sequences obtained from this study and downloaded from GenBank. They found that in model M8, 12 codons (2.3%, p> 0.95) were selected positively at a rate of dN / dS> 1, of which 8 codons (31, 24, 27, 34, 35, 38,41, 42) Corresponding to the residues of human ACE2, these codons are directly involved in the contact of human SARS virus spike proteins.
They also analyzed the ACE2 gene of R. affinis and reported that the gene occasionally binds SARS-associated coronavirus. Using the 23 ACE2 gene sequence alignments obtained from this study, it was found that the ACE2 in the entire coding region is more conservative among different individuals in the entire coding region than the ACE2 in China, and no obvious Select the site in the positive direction (data not shown).
In addition, by querying the single nucleotide polymorphism (SNP) database, they found that the SNPs in the human ACE2 gene were randomly generated throughout the coding region. Although two SNPs with non-synonymous mutations (T27A and E35K) were found in human ACE2, they all show rare frequencies (frequency of 0.00001 and 0.00002, respectively) in the global population. These results indicate that a positive selection occurs at the interface of the SARS-associated coronavirus spike protein of the bat and ACE2.