After the completion of the Human Genome Project (HGP) involving scientists from six countries, we have a preliminary grasp of the genetic “codebook”. However, genetic information is carried by cells, but until now, humans have not been able to glimpse their own cellular atlas.
Comprehensive decoding of the digital characteristics of cells will promote research in life sciences and provide basic resources and tools for the development of biomedicine. A new study led by Shenzhen BGI Life Sciences Research Institute has set its sights on macaques, which are 93% genetically similar to humans, and created a cellular map of the macaque’s whole body organs. This is also the world’s first non-human primate whole-cell map.  The study was published online on the evening of April 13 In the top academic journal “Nature”, entitled “Cell transcriptomic atlas of the non-human primate Macaca fascicularis”. The research was conducted by Shenzhen BGI Life Sciences Research Institute, Beijing BGI Life Sciences Research Institute, Shenzhen National Gene Bank, Jilin University, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, Karolinska Institutet in Sweden, University of Cambridge, Spain 35 scientific research teams from 6 countries including ICREA Research Institute and Singapore ASTAR participated in the completion.
The study was published online on the evening of April 13 In the top academic journal “Nature”, entitled “Cell transcriptomic atlas of the non-human primate Macaca fascicularis”. The research was conducted by Shenzhen BGI Life Sciences Research Institute, Beijing BGI Life Sciences Research Institute, Shenzhen National Gene Bank, Jilin University, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, Karolinska Institutet in Sweden, University of Cambridge, Spain 35 scientific research teams from 6 countries including ICREA Research Institute and Singapore ASTAR participated in the completion.
“This map is like a ‘map’, with it, it is equivalent to having a high-precision instrument for exploring the resolution of living cells, which can ‘see’ Which cells are in each organ, and the specific molecular characteristics of each cell and the interaction with other cells can be refined.” said Dr. Han Lei, the first author of the paper and Shenzhen BGI Institute of Life Sciences, ” This lays the foundation for us to better understand the basic structure of life, explore the relationship between diseases and cells, and provide a new direction for the precise treatment of diseases.”
Based on the single-cell library and sequencing platform independently developed by BGI, the research team performed single-cell sequencing analysis on about 1.14 million cells from 45 organs of adult rhesus monkeys, and divided them into 113 main cell types and 463 cell subtypes. class, and built the NHPCA (https://db.cngb.org/nhpca/) website, an interactive resource website for non-human primates millions of single cells.  One of the co-corresponding authors of the paper, Liu Longqi from Shenzhen BGI Life Sciences Research Institute, was receiving a reporter from the news (www.thepaper.cn) In the interview, he said, “From the perspective of genome sequence, the difference between macaques and humans is much smaller than that of model animals such as mice, which is the first reason for choosing monkeys; second, if you want to truly understand human diseases, such as Kinson, Alzheimer’s disease, etc., you can’t do research directly in humans, we must build animal disease models, non-human primates especially have significant advantages in cognitive and neurological disease research. “
One of the co-corresponding authors of the paper, Liu Longqi from Shenzhen BGI Life Sciences Research Institute, was receiving a reporter from the news (www.thepaper.cn) In the interview, he said, “From the perspective of genome sequence, the difference between macaques and humans is much smaller than that of model animals such as mice, which is the first reason for choosing monkeys; second, if you want to truly understand human diseases, such as Kinson, Alzheimer’s disease, etc., you can’t do research directly in humans, we must build animal disease models, non-human primates especially have significant advantages in cognitive and neurological disease research. “
Liu Longqi said that there are few such data at present, and the macaques are still blank, “so the release of these data is very important for a series of future studies. Including disease mechanism understanding, drug screening, etc. “
It is worth mentioning that, based on this map, the research team has also constructed a virus database containing 126 virus-susceptible cell types such as new crown, hepatitis B, rabies virus, etc. It’s like a “virus dictionary”, through which you can quickly look up the cell type that the virus is most likely to infect, and at the same time see the organs where the cell type is likely to be distributed.
The research team believes that with this “virus dictionary”, when doctors check the lungs of patients diagnosed with new coronary pneumonia, they may also simultaneously check the kidneys, liver and gallbladder. Because the “virus dictionary” mentions that these Organs are also distributed with cells that may be infected by the new coronavirus.In addition to diseases caused by viruses, researchers can also enter the causative genes or genetic loci of specific genetic diseases to query The likely causative cell types for the disease.
“The large-scale cellular atlas is important for our understanding of organ structural composition, embryonic development and aging, human disease, and the evolution of life. etc. are of great significance. In the future, we will also develop higher-throughput single-cell technology and multi-omics technology with spatial resolution, which will provide important tools for comprehensively building a single-cell resolution spatiotemporal map of life. Xu Xun, one of the co-corresponding authors of the paper and dean of Shenzhen BGI Life Sciences Research Institute, said, “At the same time, the cell map data is growing rapidly, and it contains a huge amount of information. The interpretation and mining of these data requires the cooperation of scientists around the world. and effort. “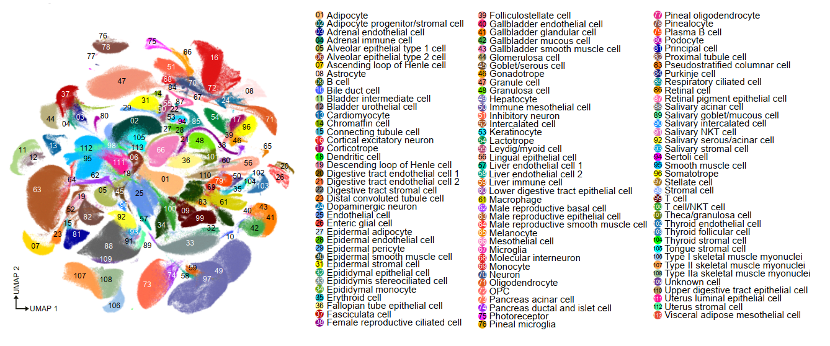
Cynomolgus monkey cell map.
It is worth mentioning that the drawing of this cell map is inseparable from the The advancement of single-cell sequencing technology and the decline in sequencing costs. Liu Longqi introduced that technology and cost constraints are also important factors affecting the progress of the Human Cell Atlas project.
The Human Cell Atlas is an international major scientific project that has emerged in the field of life sciences in recent years. It was proposed by scientists such as the United Kingdom and the United States in 2017. In the long run, the project aims to describe the detailed characteristics of each cell (about 37 trillion) in the human body , presenting the 3D structure of different types of cells in human tissues, outlining the interconnection of all human systems, and revealing the relationship between map changes and health and disease. The Human Cell Atlas project will radically improve people’s understanding, diagnosis and treatment of diseases.Liu Longqi believes that BGI was able to draw a map of macaque cells on a large scale, and the preliminary preparation was mainly due to breakthroughs in tools, “allowing us to truly open up from upstream library construction to downstream sequencing. According to the research team, at present, based on the single-cell library construction platform (DNBelab C4) and DNBSEQ sequencing technology independently developed by BGI, experts and researchers in the field can use low-cost, high-throughput, high-sensitivity and accuracy. method for large-scale single-cell sequencing analysis.
Paper link: https://www.nature.com/articles/s41586-022-04587-3